autoplot method for summary.simsum objects
Source:R/autoplot.summary.simsum.R
autoplot.summary.simsum.Rdautoplot method for summary.simsum objects
Usage
# S3 method for summary.simsum
autoplot(
object,
type = "forest",
stats = "nsim",
target = NULL,
fitted = TRUE,
scales = "fixed",
top = TRUE,
density.legend = TRUE,
zoom = 1,
zip_ci_colours = "yellow",
...
)Arguments
- object
An object of class
summary.simsum.- type
The type of the plot to be produced. Possible choices are:
forest,lolly,zip,est,se,est_ba,se_ba,est_ridge,se_ridge,est_density,se_density,est_hex,se_hex,heat,nlp, withforestbeing the default.- stats
Summary statistic to plot, defaults to
nsim(the number of replications with non-missing point estimates/SEs). Seesummary.simsum()for further details on supported summary statistics.- target
Target of summary statistic, e.g. 0 for
bias. Defaults toNULL, in which case target will be inferred.- fitted
Superimpose a fitted regression line, useful when
type= (est,se,est_ba,se_ba,est_density,se_density,est_hex,se_hex). Defaults toTRUE.- scales
Should scales be fixed (
fixed, the default), free (free), or free in one dimension (free_x,free_y)?- top
Should the legend for a nested loop plot be on the top side of the plot? Defaults to
TRUE.- density.legend
Should the legend for density and hexbin plots be included? Defaults to
TRUE.- zoom
A numeric value between 0 and 1 signalling that a zip plot should zoom on the top x% of the plot (to ease interpretation). Defaults to 1, where the whole zip plot is displayed.
- zip_ci_colours
A string with (1) a hex code to use for plotting coverage probability and its Monte Carlo confidence intervals (the default, with value
zip_ci_colours = "yellow"), (2) a string vector of two hex codes denoting optimal coverage (first element) and over/under coverage (second element) or (3) a vector of three hex codes denoting optimal coverage (first), undercoverage (second), and overcoverage (third).- ...
Not used.
Examples
data("MIsim", package = "rsimsum")
s <- rsimsum::simsum(
data = MIsim, estvarname = "b", true = 0.5, se = "se",
methodvar = "method", x = TRUE
)
#> 'ref' method was not specified, CC set as the reference
ss <- summary(s)
library(ggplot2)
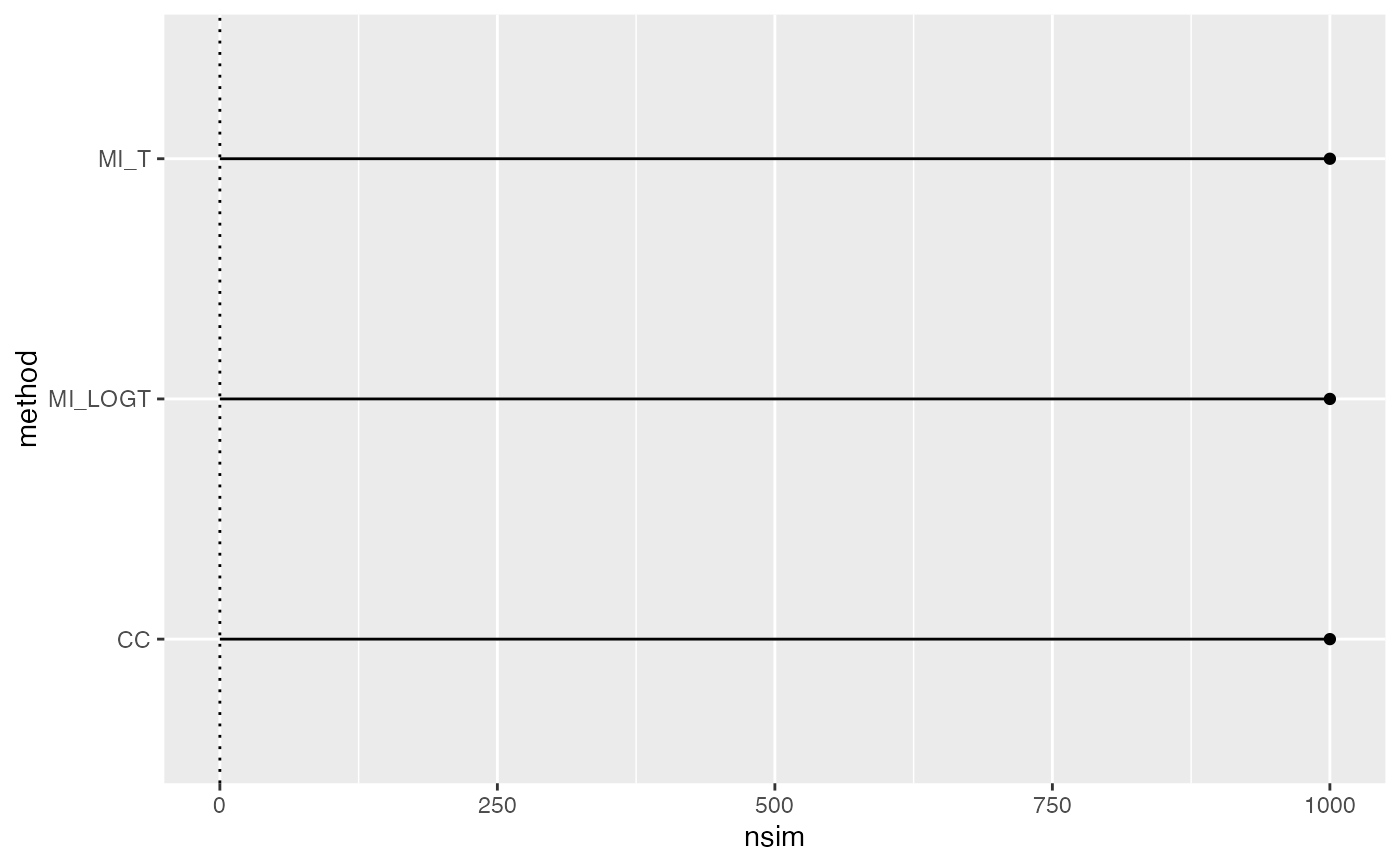
autoplot(ss)
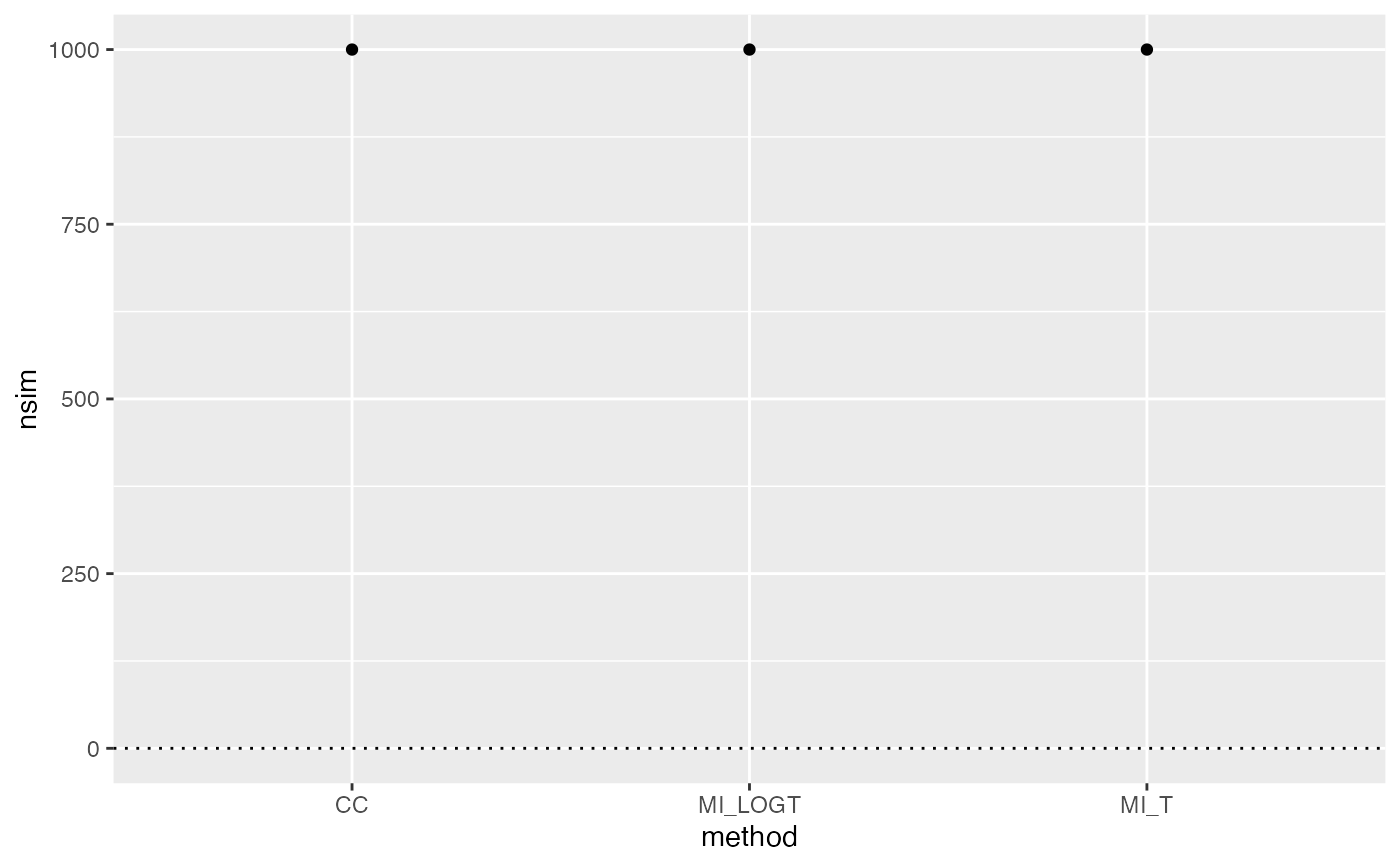 autoplot(ss, type = "lolly")
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_point()`).
autoplot(ss, type = "lolly")
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_point()`).